Reproducible research practices in magnetic resonance neuroimaging
A review informed by advanced language models
Literature overview
For this scoping review, we focused on research articles published in the journal Magnetic Resonance in Medicine (MRM). In addition to being a journal primarily dedicated to the development of MRI techniques, MRM is also at the forefront of promoting reproducible research practices. Since 2020, the journal has singled out 31 research articles that promote reproducibility in MRI, and has published a series of interviews with the article authors, discussing the tools and practicesapproaches they used to bolster the reproducibility of their findings. These interviews are freely available on the MRM Highlights portal under the label ‘reproducible research insights’.
To see how these articles relate to other literature dedicated to reproducibility in MRI, we conducted a literature search utilizing the Semantic Scholar API Fricke, 2018 with the following query terms: (code | data | open-source | github | jupyter ) & ((MRI & brain) | (MRI & neuroimaging)) & reproducib~. This search ensured that the scoping review is informed by an a priori literature search protocol that is transparent and reproducible, presenting the data in a structured way.
import requests
from thefuzz import fuzz
import json
import time
import numpy as np
import umap
import os
import pandas as pd
import umap.plot
import plotly.graph_objects as go
from plotly.offline import plot
from IPython.display import display, HTML
import base64
import plotly.io as pio
pio.renderers.default = "plotly_mimetype"
# REQUIRED CELL
DATA_ROOT = "../data/repro-mri-scoping/repro_mri_scoping"
np.seterr(divide = 'ignore')
def get_by_id(paper_id):
response = requests.post(
'https://api.semanticscholar.org/graph/v1/paper/batch',
params={'fields': 'abstract,tldr,year,embedding'},
json={"ids": [paper_id]})
if response.status_code == 200:
return response
else:
return None
def get_id(title):
"""
Query Semantic Scholar API by title.
"""
api_url = "https://api.semanticscholar.org/graph/v1/paper/search"
params = {"query": title}
response = requests.get(api_url, params=params)
if response.status_code == 200:
result = response.json()
print(result['total'])
for re in result['data']:
print(re)
if fuzz.ratio(re['title'],title) > 90:
return re['paperId']
else:
return None
else:
return None
def bulk_search(query,save_json):
"""
The returns 1000 results per query. If the total number of
hits is larger, the request should be iterated using tokens.
"""
query = "(code | data | open-source | github | jupyter ) + (('MRI' + 'brain') | (MRI + 'neuroimaging')) + reproducib~"
fields = "abstract"
url = f"http://api.semanticscholar.org/graph/v1/paper/search/bulk?query={query}&fields={fields}"
r = requests.get(url).json()
print(f"Found {r['total']} documents")
retrieved = 0
with open(save_json, "a") as file:
while True:
if "data" in r:
retrieved += len(r["data"])
print(f"Retrieved {retrieved} papers...")
for paper in r["data"]:
print(json.dumps(paper), file=file)
if "token" not in r:
break
r = requests.get(f"{url}&token={r['token']}").json()
print(f"Retrieved {retrieved} papers. DONE")
def read_json_file(file_name):
with open(file_name, 'r') as json_file:
json_list = list(json_file)
return json_list
def write_json_file(file_name, dict_content):
with open(file_name, 'w') as json_file:
json_file.write(json.dumps(dict_content))
def get_output_dir(file_name):
op_dir = "../output"
if not os.path.exists(op_dir):
os.mkdir(op_dir)
return os.path.join(op_dir,file_name)
def flatten_dict(input):
result_dict = {}
# Iterate over the list of dictionaries
for cur_dict in input:
# Iterate over key-value pairs in each dictionary
for key, value in cur_dict.items():
# If the key is not in the result dictionary, create a new list
if key not in result_dict:
result_dict[key] = []
# Append the value to the list for the current key
result_dict[key].append(value)
return result_dict
#OPTIONAL CELL
literature_records = get_output_dir("literature_records.json")
search_terms = "(code | data | open-source | github | jupyter ) + (('MRI' + 'brain') | (MRI + 'neuroimaging')) + reproducib~"
# This will save output/literature_records.json
bulk_search(search_terms,literature_records)Found 1143 documents
Retrieved 999 papers...
Retrieved 1142 papers...
Retrieved 1142 papers. DONE
1Add articles associated with the reproducibility insights¶
Among 1098 articles included in the these Semantic Scholar records, SPECTER vector embeddings Cohan et al., 2020 were available for 612 articles, representing the publicly accessible content in abstracts and titles. The high-dimensional semantic information captured by the word embeddings was visualized using the uniform manifold approximation and projection method McInnes et al., 2018.
# REQUIRED CELL
# To load THE ORIGINAL LIST, please comment in the following
lit_list = read_json_file(os.path.join(DATA_ROOT,"literature_records.json"))
# Read the LATEST literature records returned by the above search
# Note that this may include new results (i.e., new articles)
#literature_records = get_output_dir("literature_records.json")
#lit_list = read_json_file(literature_records)
# Collect all the paper IDs from the literature search
lit_ids = [json.loads(entry)['paperId'] for entry in lit_list]
# Get all paper ids for the articles linked to
insights_path = os.path.join(DATA_ROOT,"repro_insights_parsed_nov23")
insights_ids = [f.split(".")[0] for f in os.listdir(insights_path) if f.endswith('.txt')]
# Combine all IDs (unique)
paper_ids_all = list(set(lit_ids + insights_ids))
print(f"Total: {len(paper_ids_all)} papers ")Total: 1098 papers
The following cell is commented out as it involves a series of API calls that takes some time to complete.
# OPTIONAL CELL
# slices = [(0, 499), (499, 998), (998, None)]
# request_fields = 'title,venue,year,embedding,citationCount'
# results = []
# for start, end in slices:
# print(len(paper_ids_all[start:end]))
# re = requests.post(
# 'https://api.semanticscholar.org/graph/v1/paper/batch',
# params={'fields': request_fields},
# json={"ids": paper_ids_all[start:end]})
# if re.status_code == 200:
# print(f"Got results {start}:{end} interval")
# results.append(re.json())
# time.sleep(15) # Rate limiting.
# else:
# print(f"WARNING slice {start}:{end} did not return results: {re.text}")
# ALTERNATIVE
# The above API call should work fast as the requests are sent in batch.
# However, it frequently throws 429 error. If that's the case, following will
# also work, but takes much longer and a few articles may not be captured.
# results = []
# for cur_id in paper_ids_all:
# #print(len(paper_ids_all[start:end]))
# re = requests.get(
# f'https://api.semanticscholar.org/graph/v1/paper/{cur_id}',
# params={'fields': request_fields})
# if re.status_code == 200:
# results.append(re.json())
# else:
# print(f"WARNING request for {cur_id} could not return results: {re.text}")
# # Write outputs
# write_json_file(get_output_dir("literature_data.json"),results)While utilizing the Semantic Scholar API, it’s important to note that not all returned article details include word embeddings generated by SPECTER (v1) based on the title and abstract. Currently, we are filtering out articles where embeddings are available, totaling 612 out of 1098. Future efforts may focus on addressing missing data by running the SPECTER model on articles without embeddings.
# REQUIRED CELL
# Load the ORIGINAL data
lit_data = json.loads(read_json_file(os.path.join(DATA_ROOT,"literature_data.json"))[0])
# If you'd like to read from the output directory (LATEST)
#lit_data = json.loads(read_json_file(get_output_dir("literature_data.json"))[0])
papers_data = []
for res in lit_data:
if 'embedding' in res.keys():
if res['embedding']:
cur_rec = {"embedding":res['embedding']['vector'],
"title":res['title'],
"venue": res['venue'],
"year": res['year'],
"is_mrmh": "Other",
"paperId": res['paperId'],
"n_citation": res['citationCount']}
if res['paperId'] in insights_ids:
cur_rec['is_mrmh'] = "Highlights"
papers_data.append(cur_rec)
papers_data_dict = papers_data
# From a list of dicts to a dict of lists.
papers_data = flatten_dict(papers_data)
To visualize the semantic relationship between the captured articles (a 612x768 matrix), we are going to use Uniform Manifold Approximation and Projection (UMAP) method.
# REQUIRED CELL
# Reduce to 2D feature
umap_model_2d = umap.UMAP(n_neighbors=15, min_dist=0.1, n_components=2,random_state=42)
umap_2d = umap_model_2d.fit_transform(np.array(papers_data['embedding']))
umap_2d_mapper = umap_model_2d.fit(np.array(papers_data['embedding']))
# Reduce to 3D feature
umap_model_3d = umap.UMAP(n_neighbors=15, min_dist=0.1, n_components=3,random_state=42)
umap_3d = umap_model_3d.fit_transform(np.array(papers_data['embedding']))/srv/conda/envs/notebook/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning:
n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
/srv/conda/envs/notebook/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning:
n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
/srv/conda/envs/notebook/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning:
n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
Create the connectivity plot to be used as a background image for the interactive data visualization.
umap.plot.connectivity(umap_2d_mapper, edge_bundling='hammer')/srv/conda/envs/notebook/lib/python3.10/site-packages/umap/plot.py:894: UserWarning:
Hammer edge bundling is expensive for large graphs!
This may take a long time to compute!
<Axes: >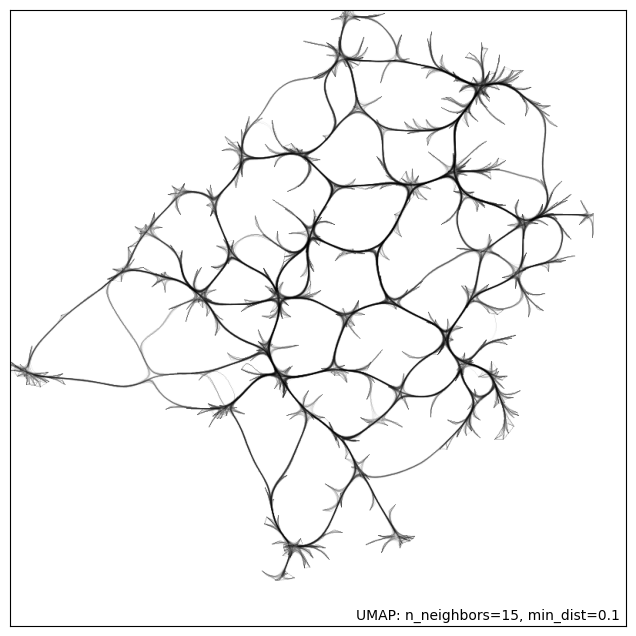
The MRI systems cluster was predominantly composed of articles published in MRM, with only two publications appearing in a different journal Adebimpe et al., 2022Tilea et al., 2009. Additionally, this cluster was sufficiently distinct from the rest of the reproducibility literature, as can be seen by the location of the dark red dots on Fig. 1.
# Create Plotly figure
fig = go.Figure()
# Scatter plot for UMAP in 2D
scatter_2d = go.Scatter(
x=umap_2d[:, 0],
y=umap_2d[:, 1],
mode='markers',
marker = dict(color =["#562135" if item == 'Highlights' else "#f8aabe" for item in papers_data['is_mrmh']],
size=9,
line= dict(color="#ff8080",width=1),
opacity=0.9),
customdata= [f"<b>{dat['title']}</b> <br>{dat['venue']} <br>Cited by: {dat['n_citation']} <br>{dat['year']}" for dat in papers_data_dict],
hovertemplate='%{customdata}',
visible = True,
name='2D'
)
fig.add_trace(scatter_2d)
# Add dropdown
fig.update_layout(
updatemenus=[
dict(
type = "buttons",
direction = "left",
buttons=list([
dict(
args=[{"showscale":True,"marker": dict(color =["#562135" if item == 'Highlights' else "#f8aabe" for item in papers_data['is_mrmh']],
size=9,
line= dict(color="#ff8080",width=1),
opacity=0.9)}],
label="Highlights",
method="restyle"
),
dict(
args=[{"marker": dict(color = np.log(papers_data['n_citation']),colorscale='Plotly3',size=9, colorbar=dict(thickness=10,title = "Citation (log)",tickvals= [0,max(papers_data['n_citation'])]))}],
label="Citation",
method="restyle"
),
dict(
args=[{"marker": dict(color = papers_data['year'],colorscale='Viridis',size=9,colorbar=dict(thickness=10, title="Year"))}],
label="Year",
method="restyle"
)
]),
pad={"r": 10, "t": 10},
showactive=True,
x=0.11,
xanchor="left",
y=0.98,
yanchor="top"
),
]
)
plotly_logo = base64.b64encode(open(os.path.join(DATA_ROOT,'sakurabg.png'), 'rb').read())
fig.update_layout(plot_bgcolor='white',
images= [dict(
source='data:image/png;base64,{}'.format(plotly_logo.decode()),
xref="paper", yref="paper",
x=0.033,
y=0.956,
sizex=0.943, sizey=1,
xanchor="left",
yanchor="top",
#sizing="stretch",
layer="below")])
fig.update_layout(yaxis={'visible': False, 'showticklabels': False})
fig.update_layout(xaxis={'visible': False, 'showticklabels': False})
# Update layout
fig.update_layout(title='Sentient Array of Knowledge Unraveling and Assessment (SAKURA)',
height = 850,
width = 884,
hovermode='closest')
fig.show()
# plot(fig, filename = 'sakura.html')
# display(HTML('sakura.html'))Figure-1: Edge-bundled connectivity of the 612 articles identified by the literature search. A notable cluster (red) is formed by the MRM articles that were featured in the reproducible research insights (purple nodes), particularly in the development of MRI methods. Notable clusters for other studies (pink) are annotated by gray circles.
fig = go.Figure()
# Scatter plot for UMAP in 3D
scatter_3d = go.Scatter3d(
x=umap_3d[:, 0],
y=umap_3d[:, 1],
z=umap_3d[:, 2],
mode='markers',
marker = dict(color =["#562135" if item == 'Highlights' else "#f8aabe" for item in papers_data['is_mrmh']],
size=9,
line= dict(color="#ff8080",width=1),
opacity=0.9),
customdata= [f"<b>{dat['title']}</b> <br>{dat['venue']} <br>Cited by: {dat['n_citation']} <br>{dat['year']}" for dat in papers_data_dict],
hovertemplate='%{customdata}',
visible = True,
name='3D'
)
fig.add_trace(scatter_3d)
fig.update_layout(
updatemenus=[
dict(
type = "buttons",
direction = "left",
buttons=list([
dict(
args=[{"marker": dict(color =["#562135" if item == 'Highlights' else "#f8aabe" for item in papers_data['is_mrmh']],
size=9,
line= dict(color="#ff8080",width=1),
opacity=0.9)}],
label="Highlights",
method="restyle"
),
dict(
args=[{"marker": dict(color = np.log(papers_data['n_citation']),colorscale='Plotly3',size=9,colorbar=dict(thickness=10,tickvals= [0,max(papers_data['n_citation'])],title="Citation"))}],
label="Citation",
method="restyle"
),
dict(
args=[{"marker": dict(color = papers_data['year'],colorscale='Viridis',size=9,colorbar=dict(thickness=10, title="Year"))}],
label="Year",
method="restyle"
)
]),
pad={"r": 10, "t": 10},
showactive=True,
x=0.11,
xanchor="left",
y=0.98,
yanchor="top"
),
]
)
# Update layout
fig.update_layout(title='UMAP 3D',
height = 900,
width = 900,
hovermode='closest',
template='plotly_dark')
fig.show()- Fricke, S. (2018). Semantic Scholar. J. Med. Libr. Assoc., 106(1), 145.
- Cohan, A., Feldman, S., Beltagy, I., Downey, D., & Weld, D. S. (2020). SPECTER: Document-level Representation Learning using Citation-informed Transformers.
- McInnes, L., Healy, J., & Melville, J. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction.
- Adebimpe, A., Bertolero, M., Dolui, S., Cieslak, M., Murtha, K., Baller, E. B., Boeve, B., Boxer, A., Butler, E. R., Cook, P., Colcombe, S., Covitz, S., Davatzikos, C., Davila, D. G., Elliott, M. A., Flounders, M. W., Franco, A. R., Gur, R. E., Gur, R. C., … Satterthwaite, T. D. (2022). ASLPrep: a platform for processing of arterial spin labeled MRI and quantification of regional brain perfusion. Nat. Methods, 19(6), 683–686.
- Tilea, B., Alberti, C., Adamsbaum, C., Armoogum, P., Oury, J. F., Cabrol, D., Sebag, G., Kalifa, G., & Garel, C. (2009). Cerebral biometry in fetal magnetic resonance imaging: new reference data. Ultrasound Obstet. Gynecol., 33(2), 173–181.